Interview with Youdiil Ophinni

Short Bio
Youdiil Ophinni became an Assistant Professor at the Hakubi Center for Advanced Research, Kyoto University, in April 2024, and joined the Center for Southeast Asian Studies as the host department. He is a visiting researcher at the Immunology Frontier Research Center, Osaka University, and the Center for Infectious Diseases, Kobe University. He earned his MD from the University of Indonesia, worked as a clinician in Indonesian hospitals, and received his PhD (Medical Science) from Kobe University in 2018. He completed his postdoctoral research at the Ragon Institute of Mass General, MIT, and Harvard in the United States during the COVID-19 pandemic. A virologist expanding into genomics and area studies, his current projects in Indonesia include a metagenomic study of viral spillover among mammals, a transdisciplinary study on the ecological impacts of the new capital in Kalimantan, and research on the intertwining issues of HIV/AIDS and illicit drug use in the country.
We are usually aware of the presence of viruses and other microbes around us only when we fall ill or experience physical ailments. Dr. Youdiil Ophinni has studied viruses and infectious diseases, beginning with HIV/AIDS in Indonesia and continuing his research in Japan and the US. This journey led him to the field of genomics—the study of genetic information in DNA to analyze the characteristics, distribution, and relationships of organisms, including the myriad invisible viruses that surround us. By leveraging the advanced technology of genomics to decipher the vast amount of information in the code of life, we may be able to respond quickly and accurately to emerging infectious diseases. Moreover, this approach may even allow us to infer the history and predict the future of living beings in a particular area, much like the methodologies used in area studies. We asked Dr. Youdiil Ophinni about his journey from virologist to genomicist, his enjoyment of research, and his ambitions for the future.
──Tell us about your research, what motivated you to pursue research, and how you came to your current research.
Being a spoiled brat as I was in my childhood, my ophthalmologist mum was exceptionally protective of my physical well-being. She would caution me that, for example, playing near parks at night could expose me to dengue-infested mosquitoes, and sharing straws with friends posed a risk of contracting hepatitis B. Even walking barefoot for wudhu (ablution before prayers) in an unclean masjid could let hookworms penetrate my soles.
Although I might not have taken her warnings seriously, and except for having dengue fever twice, I grew up to be a somewhat healthy adult (if not overweight). Nevertheless, it heightened my awareness of the countless invisible germs surrounding us, ever ready to cause illnesses unless one maintains proper immunity.


(Left) Junior high school author (age 10) in East Jakarta, 2002. (Right) During a microbiology practice in medical school at the University of Indonesia, 2007. The author (age 15) is at the center of the back row.
Born and raised in 1990s Jakarta, a rather unsanitary sprawling metropolis, my peers and I were likely exposed to myriad microbes and helminths. If the Hygiene Hypothesis holds—that frequent germ exposure strengthens the immune system—Indonesia’s Generation X would have superhuman immunity right now. During my medical school days, the subject of infectious diseases continued to fascinate me. One virus in particular intrigued me beyond belief: the human immunodeficiency virus (HIV). Instead of being gobbled up by our immune cells, this virus kills them, weakening our immunity—like a Trojan horse crippling the walls from within, or a corrupt politician sabotaging the laws of a nation.
I ended up spending the next 15 years researching HIV across multiple settings and countries. This journey as a virologist eventually led me to a new, captivating field: genomics.
The concept that life as we know it is encoded in cryptographic permutations of four letters—A, C, T (or U), and G—never ceases to amaze me. DNA and RNA govern everything, from the attachment of minuscule viruses onto cells (via the Env gene1) to how one altered gene (Tbxt2) renders mice tailless, or how a suppressed gene (ADRA2C3) heightens humans’ fight-or-flight tendency similar to chimpanzees—though without the lush body hair, due to inactive KRT41P4—and thus a propensity for conflict and war.
We used to read DNA sequences using the laborious Sanger method, akin to reading a novel letter by letter with a loupe. However, the past decade has birthed two so-called next-generation sequencing (NGS): massively parallel sequencing and ultra-long sequencing. These high-throughput approaches can decipher billions of genetic letters simultaneously, with costs plummeting every year at a pace faster than Moore’s law predicts.5 While the Human Genome Project cost one billion US dollars and took two years to complete, we can now sequence a genome for about one thousand US dollars, all in a day’s work.


(Left) Holding elution tubes after DNA extraction at Osaka University. (Right) A massively parallel sequencing device at Harvard, where I first learned about next-generation sequencing.
What can a virologist achieve with such technological prowess? In the past, we employed targeted diagnostics to identify a single virus of interest in a sample. Now, we can sequence all genomic materials—viral, host genomes, or free DNA—within a sample and match them to known libraries. Such an all-inclusive approach is known as metagenomics—it transcends a single genome.
Additionally, by comparing whole genomes of viruses side-by-side, we can trace a virus’s voyage. For instance, aligning genomes of the Coronaviridae family revealed that the first SARS virus likely jumped from cave bats in southern China to smuggled palm civets before infecting a Guangdong farmer. Similarly, the Nipah virus traveled with fruit bats displaced by deforestation to fruit orchards near piggeries in peninsular Malaysia, eventually infecting the pigs and the breeders. Thus, we can infer historical encounters between organisms based on the genomic patterns of transmitted viruses. This examination can be done even for viruses that may have altered the human genome ages ago, as pieces of viral DNA are embedded in about eight percent of our chromosomes.6


(Left) A macaque and its offspring by the roadside near Balikpapan, waiting for food from passing drivers. (Right) Together with colleagues from Airlangga University and the local zoo after a day of sampling in Surabaya.
Next-generation sequencing enables us to perform such analyses for millions of viruses across hundreds of samples simultaneously. However, I aspire to go beyond this. Microbes continuously spill over from and between living hosts, who themselves also shed genomic materials everywhere. By sequencing environmental samples—of soil, water, and the air we breathe—we can uncover a vast array of data, revealing the entire spectrum of micro- and macro-organismal interactions in a given area, including their distribution and relationships. Assembling a genomic tree allows us to see the past, tracing the ancestral evolutions of animals or the journey of hominids across the globe until they reach our area and era.7 We can even recover ancient DNA, unearthing local predecessors from thousands of years ago.8 Looking to the future, we can also project encounters influenced by climate change and other anthropogenic impacts.9
Thus, genomic analysis enables us to fathom an area’s past, present, and future, much like how anthropologists explore socio-cultural dynamics, historians delve into archival records, or political scientists scrutinize governance and power structures. Indeed, it is crucial even for virologists to adopt an interdisciplinary mindset. My goal is to integrate genomics with area studies into a new interdisciplinary field I propose to call “Area Genomics.”


(Left) A private pig farm in East Kalimantan. Nearby banana and rambutan trees may attract fruit bats, increasing the risk of microbe transmission from bat droppings to pig food. (Right) A bat population in an East Kalimantan cave. The new Indonesian capital project “Nusantara” will potentially push the bats to relocate closer to fruit plantations and animal farms.
It is a major undertaking, but I will proceed one step at a time: collecting samples from the field, extracting genomes, running them through a sequencer, and making sense of the data in an interdisciplinary context. This approach may even allow us to detect a novel pathogen before it can ravage living hosts. Naturally, I must wear protective equipment to avoid the fate of Chinese clinicians who succumbed to COVID-19 even before the disease was named. Needless to say, I also now find myself wearing flip-flops when entering masjid washrooms for wudhu.
──Please tell us about your must-have gears, or must-haves of research and writing.
When I was stationed in an infectious disease center in Surabaya, my labmates and I faced a subtle crisis. At that time, we were conducting experiments to detect hepatitis B virus (HBV) among animal sera. Strangely, we found an HBV strain known to infect gibbons also present in samples taken from orangutans and langurs. Was this evidence of cross-species HBV spillover among Old World primates? Such a discovery may have netted us a paper in an NSC journal—Nature, Science, Cell, or the pinnacle trinity for natural scientists—which, of course, was surely a pipe dream.
We shifted our attention to possible contamination. As it turned out, one of our poor gibbons had a massive HBV infection and a particularly high load of viral DNA in its blood, thus becoming a major source of contamination to other samples. The finicky thing about DNA is that it can stick to any surface, including nitrile gloves, safety cabinet walls, and, of course, pipettes. Not to mention that it is extremely durable—we can even DNA-type a 15-year-old blood stain. RNA is the polar opposite; one unmasked cough means goodbye to the RNA I am holding at my pipette tip.

In any case, it was fortunate that I brought my trusted pipettes, which I used to conduct a conclusive round of tests and clear up the misunderstanding. I purchased them right after I earned my PhD and obtained my first research grant. Finally free to use “my own” money, and growing tired of thumb spasms from the old Gilson pipettes my lab boss had, I splurged on the higher-end Eppendorf brand. I treasured them and did not use them much, but I happened to carry them to each lab I relocated to and, gratefully, they saved my work much later.
Of course, I don’t bring them to the field—no virologist in their right mind would take their pipettes out of the lab (although I did exactly that). For that, my trusted gear during frequent flights includes my headphones, a good book or two, and my portable gaming system.
──Tell us about the difficulties and ingenuity you faced while conducting your research, or compiling your research results into a paper or book.
For the past decade, the trend in molecular biology research has been the “omics” approach, which aims to collectively characterize the entirety of a given biological object. This object is denoted by the suffix “-ome.” For example, I focus on the genome (genes or DNA) and virome (viruses), while others may investigate the proteome (proteins) and transcriptome (RNA transcripts). These approaches attempt to collect all possible data from samples without a specific target in mind—in other words, blindly (or unbiasedly is the preferable term). Counterintuitive to the customary conception of research questions, we exhaustively gather data first, then seek patterns, and finally formulate questions based on the findings.
Omics methods have faced criticisms, particularly from traditional, hypothesis-driven biologists.10 These approaches generate excessive “junk” data, are prone to false discoveries, face technical and computational bottlenecks, and often lack reproducibility. Indeed, omics produces massive amounts of data, quickly filling terabytes of storage. Indeed, I experienced one of the most traumatic moments in my career when my hard drive containing metaviromic data abruptly stopped spinning (teaching me to always invest in SSDs).
Interpreting this data, to find glimpses of eureka amid the sea of noise, is a daunting task. Even afterward, validation is vital. For virome studies, this entails old-school wet lab experiments to confirm the presence of a virus by isolating it and demonstrating its signs of life: the ability to replicate and adapt (or mutate). This is no easy task; I am often reminded of what my senpai, Tomohiro Kotaki-sensei (now at the Research Institute of Microbial Disease (RIMD) at Osaka University), used to say when I was a greenhorn: “Ninety-nine percent of experiments are bound to fail.” But, to ensure that we not methodologically “mess up” the one percent, we must follow the protocol one hundred percent of the time.
The greatest challenge, I believe, lies in determining the biological relevance. Even when a discovery is made, how is its significance and “disruptive”11 capacity within the broader context of biology? Does it advance our understanding of life? Finally, it is just a matter of writing a coherent report and defending our arguments to the hard-to-please peer reviewers.
──Could you comment on what books you would recommend to young people?
It may sound cliché to suggest reading books outside our own disciplines. I would do that in a heartbeat—reading should be entertaining, and it feels dreary to read further into your own field (or scientific journals, for that matter) outside of work hours. I remember being excited when reading Ibn Khaldun’s historiography, Greene’s physics, Said’s philosophy, and Frenkel’s mathematics, as well as fiction books, from Dostoevsky, Mishima, Asimov, and Pramoedya to Dan Brown and Ishiguro. Inspirations for choosing my field, however, were Oldstone’s Viruses, Plague, and History, Dawkins’s The Selfish Gene, and Mukherjee’s The Gene: An Intimate History. My current readings are also closer to work: Frans de Waal’s books on primate behavior and evolution.
I prefer to suggest that young scholars join a reading club. Engaging with diverse minds fosters out-of-the-box thinking, exposes nuances we may have missed, intellectually challenges our interpretations, or, at the very least, provides a sounding board for the ideas we have gathered from the book. I joined one immediately after coming to CSEAS, organized by Professor Michael Feener, and I wish I had done so when I was younger.
──Please say a word to those aspiring to become researchers.
To be a researcher, having curiosity is a given. However, one often overlooked trait, I believe, is humility. It is essential to remain humble and acknowledge when we do not know something. By setting aside ego and embracing the mentality of a naïve child, we can absorb information like a sponge and avoid the Dunning-Kruger effect. We have a saying in Bahasa: “Be like a rice stalk; the more it is filled with ripening grains, the more it bows down.” I tend to sense humility among Japanese researchers rather than Indonesians, however.
I think that is one of the benefits of interdisciplinary research. The more I engage with academics from other fields, the more it reveals the limits of my own knowledge, and, quite frankly, just how dumb I am.
──Tell us about your ambitions for the future.
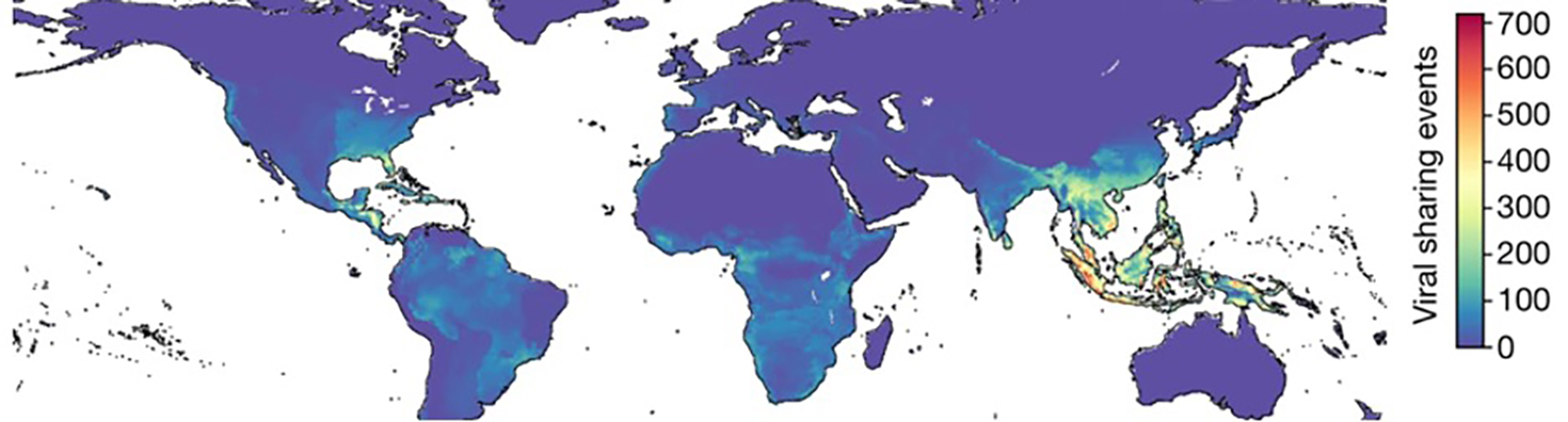
The map above illustrates the prediction of cross-species viral spillover driven by climate change.9 Southeast Asia faces the highest risk, yet countries like Indonesia and the Philippines lack robust surveillance systems for early detection of emerging infectious diseases. The preparedness of virology centers in Wuhan and the Global North is truly enviable—consider how swiftly they sequenced SARS-CoV-2, the virus responsible for COVID-19, and mapped even minute mutations in its progeny strains. It is imperative that all Southeast Asian countries develop similar high-throughput sequencing capabilities, operating around the clock for rapid, evidence-based policymaking. I dream of such a prospect for my home country.
I once learned of a virologist friend in the UK who ingeniously connected power to his portable DNA sequencing device using a dynamo attached to his bicycle. Every time he got a new genetic sample from the field, he would pop it into his sequencer and run the machine while cycling back across town to his lab. Having recently learned how to bike myself, I would love to do so too.

References
- Arrildt, K. T., Joseph, S. B. & Swanstrom, R. The HIV-1 Env Protein: A Coat of Many Colors. Curr. HIV/AIDS Rep. 9, 52–63 (2012). ↩︎
- Xia, B. et al. On the genetic basis of tail-loss evolution in humans and apes. Nature 626, 1042–1048 (2024). ↩︎
- Lee, K. S. et al. Selection on the regulation of sympathetic nervous activity in humans and chimpanzees. PLOS Genet. 14, e1007311 (2018). ↩︎
- Winter, H. et al. Human type I hair keratin pseudogene ϕhHaA has functional orthologs in the chimpanzee and gorilla: evidence for recent inactivation of the human gene after the Pan-Homo divergence. Hum. Genet. 108, 37–42 (2001). ↩︎
- DNA Sequencing Costs: Data. https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data. ↩︎
- Ophinni, Y., Palatini, U., Hayashi, Y. & Parrish, N. F. piRNA-Guided CRISPR-like Immunity in Eukaryotes. Trends Immunol. 40, 998–1010 (2019). ↩︎
- Pedro, N. et al. Papuan mitochondrial genomes and the settlement of Sahul. J. Hum. Genet. 65, 875–887 (2020). ↩︎
- Mühlemann, B. et al. Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature 557, 418–423 (2018). ↩︎
- Carlson, C. J. et al. Climate change increases cross-species viral transmission risk. Nature 607, 555–562 (2022). ↩︎
- Nurse, P. Biology must generate ideas as well as data. Nature 597, 305–305 (2021). ↩︎
- Park, M., Leahey, E. & Funk, R. J. Papers and patents are becoming less disruptive over time. Nature 613, 138–144 (2023). ↩︎
This article is also available in Japanese. >>
「生命の暗号的足跡:地域ゲノミクスによる生物間の遭遇の解読」
オフィンニ ユディルさんインタビュー